
Application Note
Imaging characterization of a 3D bioprinted ovarian cancer model
- The combination of high-content screening and transient transfection helps characterize and validate 3D bioprinted assays
- Transient gene expression is a powerful tool to increase the versatility of 3D bioprinted models
- Multi-wavelength analysis tools are essential to fully exploit the power of 3D bioprinted models to study the interaction of different cell type populations within the model
Z. Baka, C. Godier, H. Alem | Institut Jean Lamour
R. Storm, M. Cattaneo, S. Tendil | Molecular Devices
N. Chaboche, A. Nyamay’Antu | Polyplus Transfection
V. Gribova | INSERM
Introduction
3D bioprinting is defined as the additive deposition of cells and biocompatible materials to build biologically functional 3D structure or artificial tissue models.1 This technology has revolutionized the field of tissue engineering, making it possible to create structures with a complex, predefined design while ensuring good reproducibility. Several 3D bioprinting-based approaches have been reported for engineering various tissues such as cartilage,2 bone,3 as well as for skin regeneration.4 This technology is also exploited to create new preclinical cancer models. Indeed, as 2D cell culture-based models are increasingly questioned for their lack of predictivity, 3D bioprinting has emerged as an alternative approach to circumvent this problem by deposing different cell types and extracellular matrix derived molecules for better modelling/representation of the tumor microenvironment (TME). Many 3D bioprinting-based cancer models have been reported, including lung cancer,5 breast cancer,6 and glioblastomas.7 These models show advantages in design flexibility and reproducibility, compared to other strategies such as spheroids and organoids.
3D bioprinting is still under investigated as a model of ovarian cancer which is a major public health issue. In this type of tumor, cancer associated fibroblasts (CAFs) were shown to closely interact with cancer cells and play a major role in cancer invasion and drug resistance.8,9 CAFs are thus a key component to consider when designing an ovarian cancer model.
In this application note, 3D bioprinting is used to create an ovarian cancer model comprising of cancer cells (SKOV3 cells) and CAFs fibroblasts (MeWo cells). The SKOV3 cells were transfected with a GFP plasmid using jetOPTIMUS® (Polyplus®) and the MeWo cells were stained with CellTracker™ Orange CMRA Dye (ThermoFisher). The two cell types were included in a gelatin-alginatebased hydrogel to obtain a “bio-ink” that was used to create cylindrical tumor-like structures. Homogeneity and reproducibility of cell distribution within these tumor models, were assessed using the ImageXpress® Pico imaging system.
Results and discussion
As shown in Figure 1, imaging of bioprinted structures show that GFP expressing SKOV3 cells and fluorescently dyed MeWo cells are homogenously distributed within the gelatin-alginate-based matrix (Figure 1A and B). This is important as homogenous cell distribution within the bioink is a crucial parameter to be considered while performing 3D bioprinting.12 Moreover, the native TME is known to comprise several cell types that closely interact with each other.13 Therefore, it is crucial to reproduce this cell proximity while building an in vitro tumor model to promote interactions between cancer and stroma cells.
While many cell types should be combined in a tumor model to better mimic the TME heterogeneity, the total number of each cell type and their respective proportions need to be controlled to ensure reproducibility. The ImageXpress Pico is a particularly suitable system to perform such controls with its ability to quickly and easily capture multiple microscope images. Its software Cell ReporterXpress allows the quantification of multiple cell populations even in a thick sample using the Z-stack maximum projection. In this application note, this system was used to analyze our bioprinted tumor models which showed good reproducibility in terms of cell number and proportions. Among the 6 considered bioprinted structures, the average number of SKOV3 cells (cell count FITC) was 837 ± 200 (standard deviation ≤ 25%). For MeWo cells (cell count TRITC), the average number was 3264.50 ± 462 (StD ≤ 15%).
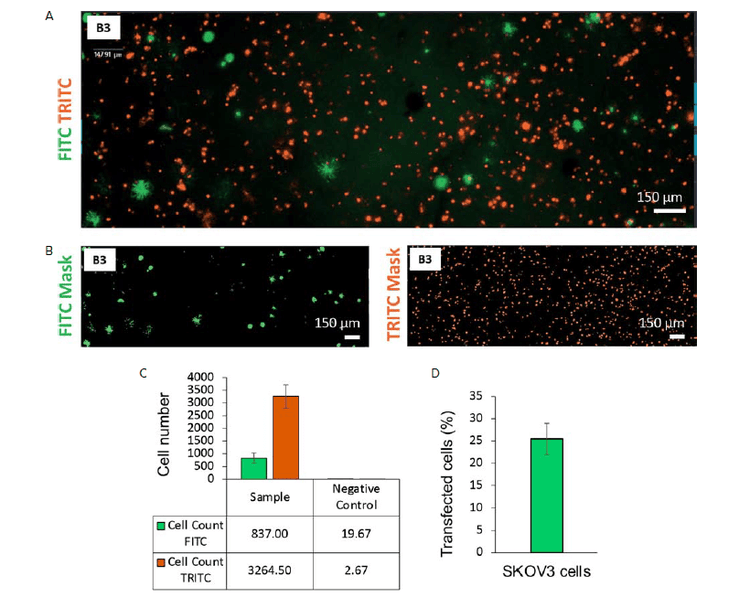
Figure 1. Analysis of a 3D bioprinted structure containing SKOV3 cells transfected with a GFP plasmid and MeWO cells stained with CellTracker Orange CMRA Dye. A) FITC and TRITC images of the 3D bioprinted structure in well B3 acquired with the ImageXpress Pico system (4X, Z-stacking). B) Analysis mask for FITC and TRITC cell count created using CellReporterXpress software. C) Average cell numbers in FITC and TRITC channels for sample (5 wells, stained and transfected) and negative control (3 wells, unstained and not transfected). D) Estimation of the transfection efficency expressed as average percentage of transfected cells using GFP plasmid and jetOPTIMUS-Polyplus-transfection-transfection kit (% of FITC positive cells / TRITC positive cells).
This application note demonstrates how the visualization of specific genes can quickly be observed in bioprinted structures. Transient gene expression can be achieved in specific cell types of the bioprinted structure by transfecting them before incorporating them in 3D constructs via bioprinting. This is a significant advantage of 3D bioprinting over other 3D cell culture methods such as spheroids and organoids where transfection is known to be challenging. In this preliminary experiment, the estimation of transfection efficiency is 25% (StD 3.48%) (Figure 1D). This could be improved, for instance, by using the ImageXpress Pico to screen multiple concentrations of transfection mix, DNA plasmid amount and timing. This approach also provides the possibility to perform gene silencing and or gene editing in both stroma and cancer cells within the same bio-printed structure.
Overall, we combined transfection, fluorescent imaging and cell counting to characterize and validate a 3D bioprinted ovarian tumor model containing cancer cells and CAFs. We could demonstrate reproducible and homogenous distribution of the two cell types and their proximity in the matrix which is favorable for cell-cell interactions. This approach offers the possibility to create more complex and complete tumor models with controlled and reproducible cell distributions, and without the limitations of dye penetration observed in 3D models. 3D bioprinted tumor models can therefore be rapidly validated for their reproducibility before integration into microfluidic systems. They can then be cultured longterm under physiological flow to create more in vivo-like preclinical models for anticancer drug development.
Materials and methods
Hydrogel preparation
For hydrogel preparation, the required masses of gelatin and sodium alginate powders were weighed, decontaminated under UV rays for 1 hour, then dissolved in Dulbecco’s Modified Eagle Medium mixed with Minimum Essential Medium (MEM) supplemented with 10% fetal bovine serum. The obtained solution was then kept at 37°C under magnetic stirring overnight for complete homogenization.
Cell transfection and staining
SKOV3 cells were transfected with a GFP plasmid using jetOPTIMUS (Polyplus) according to the manufacturer’s instructions.10 Briefly, the appropriate amount of DNA was diluted into the jetOPTIMUS buffer and the appropriate volume of jetOPTIMUS transfection reagent was added. The transfection solution was kept at room temperature for 10 minutes before being added to SKOV3 cells cultured in T75 flask at 60–80% confluence.
MeWO cells were stained with CellTracker Orange CMRA Dye (ThermoFisher) according to the manufacturer’s instructions.11 Briefly, a working solution was prepared by dissolving the dye in DMSO and then diluted in MEM culture medium. Cells were exposed for 30 minutes at 37°C. The reagent solution was then removed and replaced by fresh complete culture medium.
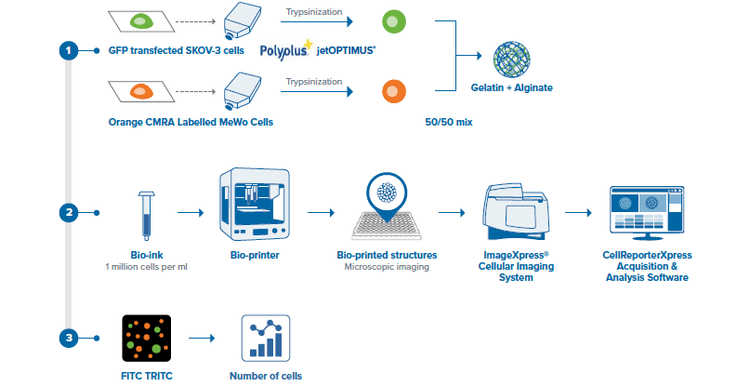
Figure 2. Schematic description of the method used to bioprint and image the ovarian cancer model structures
Cell encapsulation
For bio-ink preparation, trypsinized cells were resuspended in fresh culture medium and carefully added to the previously prepared polymer solution. The obtained suspension was slowly agitated to homogenize cell distribution within the bio-ink.
Bioprinting process
The prepared bio-ink was loaded in a 3 mL cartridge and loaded into in the bioprinting head. Bioprinting was then performed in 24-well plates. Each 3 mL cartridge bioprinted about 48 structures. After bioprinting, the structures were crosslinked using a 100 mM CaCl2 solution, supplemented with fresh culture medium and cultured at 37°C and 5% CO2 until required for experimentations.
Cell imaging
Bioprinted structures were transferred into a 96 well plate (Greiner 655892, glass bottom, black wall).
Cells were imaged with the ImageXpress Pico Automated Cell Imaging System using the 4X objective.
Images were acquired at one site per well with transmitted light, FITC and TRITC channels at 5, 150, and 250ms exposure times, respectively. A 4X objective displays around 40% of the total well area. A combination of Hardware-based and Image-based autofocus was used to acquire seven z-planes with 50 µm separation (Z Stacking). Maximum 2D projection images were generated on-the-fly
The preconfigured cell count analysis module in Cell ReporterXpress™ software was used to quantify cells with a signal either in the FITC or TRITC channels. The segmentation parameters used for the analysis were Intensity, Minimum and Maximum width. The values were 23, 7, 100 for the FITC channel and 23, 7, 38 for the TRITC channel.
Acknowledgments
Pascal Kessler – Microscopy platform of CRBS PIC-STRA, Strasbourg, France.
References
- Tan B, Gan S, Wang X, Liu W, Li X. Applications of 3D bioprinting in tissue engineering: advantages, deficiencies, improvements, and future perspectives. J Mater Chem B. 2021;9(27):5385–5413. doi:10.1039/D1TB00172H
- Nguyen LH, Kudva AK, Saxena NS, Roy K. Engineering articular cartilage with spatially-varying matrix composition and mechanical properties from a single stem cell population using a multi-layered hydrogel. Biomaterials. 2011;32(29):6946–6952. doi:10.1016/j. biomaterials.2011.06.014
- Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat Biotechnol. 2016;34(3):312–319. doi:10.1038/nbt.3413
- Cubo N, Garcia M, Cañizo JF del, Velasco D, Jorcano JL. 3D bioprinting of functional human skin: production and in vivo analysis. Biofabrication. 2016;9(1):015006. doi:10.1088/1758-5090/9/1/015006
- Wang X, Zhang X, Dai X, et al. Tumor-like lung cancer model based on 3D bioprinting. 3 Biotech. 2018;8(12). doi:10.1007/s13205-018-1519-1
- Swaminathan S, Hamid Q, Sun W, Clyne AM. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication. 2019;11(2):025003. doi:10.1088/1758-5090/aafc49
- Wang X, Li X, Ding J, et al. 3D bioprinted glioma microenvironment for glioma vascularization. J Biomed Mater Res B. Published online 2020. doi:10.1002/jbm.a.37082
- Ji Z, Tian W, Gao W, Zang R, Wang H, Yang G. Cancer-Associated Fibroblast-Derived Interleukin-8 Promotes Ovarian Cancer Cell Stemness and Malignancy Through the Notch3-Mediated Signaling. Front Cell Dev Biol. 2021;9:684505. doi:10.3389/fcell.2021.684505
- Yan H, Guo BY, Zhang S. Cancer-associated fibroblasts attenuate Cisplatin-induced apoptosis in ovarian cancer cells by promoting STAT3 signaling. Biochem Biophysl ResCommun. 2016;470(4): 947–954. doi:10.1016/j.bbrc.2016.01.131
- jetOPTIMUS® DNA Transfection Reagent. Accessed June 9, 2022. (account creation required) https://myaccount.polyplus-transfection.com/my-account-home/protocol-download/
- CellTrackerTM Orange CMRA Dye. Accessed June 8, 2022. https://www.thermofisher.com/order/catalog/product/C34551
- Bhattacharyya A, Janarthanan G, Tran HN, Ham HJ, Yoon J, Noh I. Bioink homogeneity control during 3D bioprinting of multicomponent micro/nanocomposite hydrogel for even tissue regeneration using novel twin screw extrusion system. Chem Eng J. 2021;415:128971. doi:10.1016/j.cej.2021.128971
- Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30(16):R921–R925. doi:10.1016/j.cub.2020.06.081