
Application Note
Enhanced development of virus-specific hybridomas using ClonePix and CloneSelect Imager Technologies
- Screen more clones than traditional methods to increase the probability of finding rare, high secretors
- Unique process produces hybridomas with increased titer levels
- Increase probability of finding optimal producers
Introduction
Double-stranded DNA (ds-DNA) viruses cause a variety of disease in humans. Species within the order of Herpesvirus and the families Adenovirus and Papillomavirus can cause symptoms in humans with similar presentations. Furthermore, serological and protein-based diagnostic confirmation for a particular virus may be complicated due to antigen conservation within viral genomes. A high degree of proteomic homology and serum cross reactivities lead to poor differentiation between individual species of ds-DNA viruses. The purpose of the research was to identify an optimal producer cell line that secrets a highly specific, non-cross-reactive monoclonal antibody to be used in biotherapeutics development.
The ClonePix™ System was employed in the highthroughput screening of hundreds of sub-cloned colonies from parental hybridoma material (historically parental line yields were less than 1 mg/L of mAb). Viable, highIgG secreting clones were further objectively assessed for growth characteristics using CloneSelect™ Imager technology. This highly efficient automated clone selection aided rescue and stabilization of a high-titer hybridoma cell line producing a highly specific antibody to an immunogenic viral antigen. The pairing of technologies provided sufficient material for immunodiagnostic device development.
Materials
- ClonePix System (Molecular Devices, LLC)
- CloneSelect Imager (Molecular Devices, LLC)
- CloneMedia Semi-Solid Media for hybridomas/myelomas (Molecular Devices, LLC Cat #K8600/K8610)
- CloneDetect Reagent, Mouse IgG (H+L) Specific, Fluorescein (Molecular Devices, LLC Cat #K8220).
Rescuing poorly producing hybridomas
Traditional methods for hybridoma development involve chemically-mediated fusion of a myeloma cell line with the splenocytes of a host animal immunized with an antigen of interest. The fused hybridomas undergo notoriously slow and inefficient selection processes by limiting dilution, followed by ELISA screening to isolate a limited number of antigen-specific monoclonal hybridoma lines. With an extended timeline (up to 30 weeks) and increased handling, limiting dilution methods produce lower titer cell lines where the vitality and productivity of the cell line could never be precisely elucidated.
The first ds-DNA viral specific hybridoma cell line was developed in 1990s using limiting dilution methods, thus the monoclonality and stability was not optimized. Repeated cultures (static and roller) were done over the years with varying sera and media compositions, yet the clones were rendered unstable (Figure 1). Moreover, antibody yields never reached more than 3 mg/L range and the quality of the purified IgG was never uniform with varying amounts of aggregation observed in the preps.
The goal was to rescue and stabilize the specific hybridoma cell line by sub-cloning a higher titer antibody producing parental line. ClonePix Platform offers a significant technological advantage utilizing a semisolid methylcellulose media and label-free detection by CloneDetect Reagent to rescreen a very large number of previously fused hybridomas (Figure 2).
High throughput screening methodology using a ClonePix System enabled the rescue of stable, higher producing, stable clones from the lower producing parental clones.
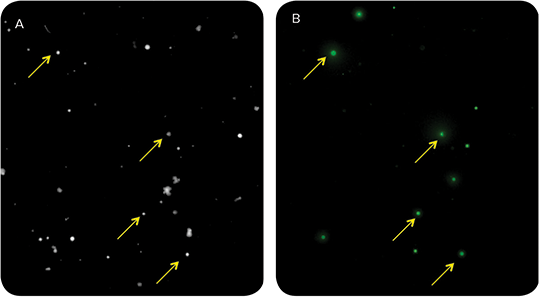
Figure 1. Visualization of IgG secretion in parental hybridoma cell line suggests only a small subset (~7%) is producing ds-DNA virus-specific antibody. Hybridoma cells were plated at 200 cells/mL in 6-well plates in CloneMedia Semi-Solid Media (for hybridomas/myelomas). Fluorescently conjugated CloneDetect Agent was added to enable in situ detection of secreted IgG. Colonies were imaged on a ClonePix System in white light at 150 ms (A) and fluorescence at 500 ms (B). Variability of FITC signal as quantitated by Interior Mean Intensity and Exterior Mean Intensity of the parent line indicate an unstable, non-monoclonal hybridoma cell line.

Figure 2. ClonePix Technology Principle. A: Concept overview of antibody detection from individual hybridoma clones grown suspended in a methylcellulose matrix. B: Whitelight image of precipitated antibody sequestered around a growing hybridoma clone. C: ClonePix System detection and selection of acceptable target clones. D: ClonePix System automated picking and transfer of isolated target clones to destination plates.
Rapid isolation of highyielding target clones secreting antibody
ClonePix Systems have demonstrated significant improvements in workflow productivity and costeffectiveness, increasing the probability of finding rare secretors, thereby reducing the time required for monoclonal antibody generation by up to 50% as compared to limiting dilution methods.
In this process, parental hybridoma clones were expanded, followed by analysis and picking of the desired clones on ClonePix Systems. Using the FITC-labeled CloneDetect Reagent, clones were imaged and analyzed for fluorescent intensity using the ClonePix Software. FITC positive clones were ranked and picked based on the high exterior mean intensity [FITC] values. Overall, 480 FITC-IgG positive clones were picked which represent only 5–6% of the total population of parental clones screened. The majority of high secreting clones were small (40–60 cells) versus the larger, faster growing clones (70–150 cells) that were determined to be negative IgG secretors (Figure 3).

Figure 3. Detection and selection of viable, high secreting colonies using ClonePix Software analysis. A: Scatter plot scatter plot shows a liner correlation between Exterior and Interior Mean Intensity suggesting that the IgG is being secreted properly (otherwise immobilized on the cell surface with high Interior and low Exterior), and the low IgG yielding clones are due to a heterogenous population of variable secretors (i.e. only a few hybridomas, 5–6% of the total population, are producing IgGs, while the majority of the clones are growing without IgG expression). B: Ranking plot shows clone selections in purple: the cutoff was FITC signals to pick 480 clones (Exterior above 700 and Interior above 1000).
Four hundred and eighty high secretors were picked from source plates with semi-solid CloneMedia and deposited into five 96-well destination plates containing 200 µL of liquid cloning media, then allowed to expand for three days. CloneSelect Imager was used to image picked clones in 96-well plates in order to objectively and quantitatively assess cell growth. Superior imaging speed (less than 90 seconds for 96-well plate) enabled to rapidly monitor and evaluate colony growth for five days. By effectively utilizing CloneSelect Imager process, 146 top growers were isolated (Figure 4).
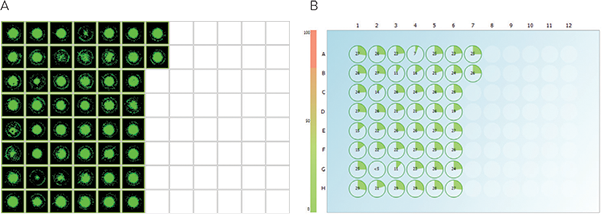
Figure 4. CloneSelect Imager Software analysis enables identification of optimally growing clones. Image analysis of the subset of 480 picks demonstrates assessment of colonies’ cellular morphology and growth characteristics (A) as well as estimation of colony number and colony area for each well (B) thereby efficiently tracking colony growth. Slower growing colonies were omitted, while the top growers—30% of the total population (146 colonies)—were used to screen for specificity with peptide assay.
Screening and identification of sub-clones for pilot scale analysis
One hundred forty-six top growers were screened for antigen specificity with a synthesized peptide representing the epitope for mAb binding. The peptide had exhibited a strong affinity for antibody as demonstrated by previous studies and thereby was used to screen novel subclones secreting high affinity antibodies.
Biotinylated peptide was loaded onto streptavidin surface followed by the addition of undiluted hybridoma supernatants from 146 samples to measure for IgG binding. Top seven highly specific sub-clones had exhibited optimal binding and were chosen for banking.
Pilot scale productions of high IgG secreting sub-clones
Seven high-valued, specific sub-clones were scaled up in liquid culture for future analysis. They were expanded and once sufficient volumes of culture were obtained, five sub-clones were frozen for future reference studies, while the remaining two high binding/high IgG secreting clones were expanded for 2–3 weeks and pilot scale productions (1 Liter) were completed.
Workflow to rescue poorly producing hybridomas
To identify sub-clones for advancing diagnostic development for ds-DNA virus-specific antibody, 480 FITC positive hybridoma clones were screened using ClonePix Technology, including CloneMatrix Semi-Solid Media and FITClabeled CloneDetect Reagent. Of the 146 fastest growing clones as identified by CloneSelect Imager, seven high producers were determined to be highly specific for target peptide binding. Two subclones were selected for pilot production and cell banking. This workflow reduces hybridoma screening time by up to 50%, while selecting robust, desired candidates for additional research or production.
Verification of monoclonality and uniform IgG secretion
After ClonePix System recloning and confirmation of specificity, two subclones were expanded and re-plated at 200 cells/mL in CloneMedia Semi-Solid Media in 6-well plates with CloneDetect Reagent. Visualization and analysis by the ClonePix System software suggested that IgG was being secreted properly and higher IgG yields were observed due to a homogeneous population of uniform secretors (Figure 5).
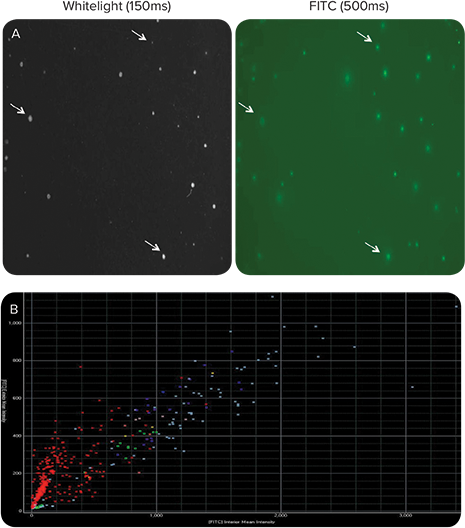
Figure 5. Sub-cloning of parental hybridoma by ClonePix results in uniform IgG secretion at higher yields. A subset data for one sub-clone is shown. A: A significant improvement in % FITC-positive colonies as a result of sub-cloning is observed as compared to the parental clone (Figure 1 A, B). IgG secretion was detected at seven days growth by 100 U/mL of CloneDetect Reagent. B: Scatter plot shows a linear correlation between Exterior and Interior Mean Intensity; the slope is shifted towards the Y-axis indicating greater uniformity, while more clones in upper quadrants signify the presence of higher FITC positive clones.
Production capacity on novel ds-DNA viral hybridoma sub-clones
By utilizing ClonePix Technology poor producing parental clones were re-screened and higher producing, stable sub-clones were rescued. mAb productions were purified over a protein G column and total yields quantified. Both novel sub-clones had shown a dramatic improvement in IgG production (17-25 mg/L) over historic yields of the parent clone (\~1mg/L), shown in Figure 6.
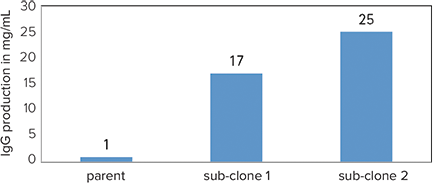
Figure 6. ds-DNA virus IgG clone production capacity. Sub-cloning by ClonePix System resulted in a dramatic increase in cell line titer.
Conclusion
ClonePix Technology improves both the timeline and quality of screened hybridoma sub-clones by decreasing manual manipulation, while CloneMatrix Media and CloneDetect Reagent allow improved growth and high sensitivity detection of secreting clones. Data from the ClonePix System suggests that the poor production capacity of the parent clone was due to a small subpopulation of antibody producing cells that were being overtaken by faster growing, non-producing cells.
Used in conjunction with CloneSelect Imager, the ClonePix System had proven to be a powerful tool in identifying two high-yield, high-affinity hybridoma subclones capable of significantly higher antibody production capacity (17-25 mg/L) as compared to the previous production attempts of the parental line hybridomas (0.5-1 mg/L). Thus, this technology effectively contributes to the development of a novel therapeutic antibody to detect specific ds-DNA virus.