
Application Note
Analysis of apoptosis using automated cell imaging
- Evaluate apoptosis using a simplified, no-wash, homogenous assay
- Acquire and analyze images on the fly to get to your results faster
- Monitor cell health and quantify apoptosis using preconfigured analysis modules
Introduction
Matthew Hammer and Oksana Sirenko, PhD | Applications Scientists | Molecular Devices
Apoptosis is a process of programmed cell death that occurs in multicellular organisms.1,2 Biochemical events lead to characteristic changes in cell morphology and cell death. Morphological changes include cell shrinkage, nuclear fragmentation, chromatin condensation, chromosomal DNA fragmentation, and mRNA decay. Apoptosis is a highly regulated process that can be initiated via the intrinsic pathway as a response to various stressors including starvation, infection, hypoxia, oxidative stress, etc. Mitochondria damage plays an important role in the initiation of apoptosis. In the extrinsic pathway, apoptosis is initiated by external signals, including tumor necrosis factor receptor family-mediated signaling. Both pathways induce cell death by activating caspase enzymes. Disruptions in the regulation of apoptosis have been implicated in various diseases, including cancer.3 Here, we describe a cell-based assay for apoptosis detection using automated cell imaging.
Materials
- HeLa cells (ATCC P/N CCL-2)
- HeLa media
- DMEM: CellGro, with L-glutmaine (Corning)
- 10% FBS (BenchMark™; Gemini P/N 100-106)
- 1% penicillin/streptomycin
- Staurosporine (Sigma P/N S5921)
- Mitomycin C (Sigma P/N M4287)
- Camptothecin (Sigma P/N C9911)
- EarlyTox Caspase-3/7 NucView 488 Assay Kit (Molecular Devices P/N R8348)
- 96-well black, clear-bottom microplates (Greiner P/N 655090)
- ImageXpress Pico Automated Cell Imaging System and CellReporterXpress Automated Imaging and Analysis Software

Method
HeLa cells, a human cervical carcinoma cell line, were plated in black, clear-bottom, 96-well Greiner microplates at 5,000 cells per well and were grown overnight at 37°C, 5% CO2. The following day, the cells were treated in quadruplicate with different concentrations of anti-cancer compounds—staurosporine, mitomycin C, or camptothecin—to induce apoptosis. The highest concentrations of each compound were 10 µM staurosporine, 200 µM mitomycin C, and 100 µM camptothecin. At 18 hour post treatment, the cells were stained for 30 minutes with EarlyTox™ Caspase-3/7 NucView 488 dye and Ethidium Homodimer III, for final 1x concentrations of 5 µM and 3 µM, respectively. Hoechst 33342 nuclear dye was then added to each well for a final concentration of 6 µM, and the cells were placed back into the 37°C, 5% CO2 incubator for 15 minutes.
EarlyTox Caspase-3/7 NucView 488 Assay Kit
Caspase-3 and caspase-7 are proteases that are activated during the execution phase of apoptosis. Caspase-3/7 NucView 488 substrate from the EarlyTox kit is used to detect caspase-3/7 activity within intact cells. The substrate consists of a fluorogenic DNA dye that is coupled to the caspase-3/7 DEVD recognition sequence. Initially nonfluorescent, the substrate permeates the plasma membrane and enters the cytoplasm. In apoptotic cells, caspase-3/7 cleaves the substrate, releasing a high-affinity DNA dye that migrates to the cell nucleus and binds to DNA. Excitation at 500 nm results in an emission of a bright green fluorescence at 530 nm and can be used to evaluate the level of caspase activation. Hoechst 33342 nuclear dye was used for evaluation of the total cell number and Ethidium Homodimer for the number of dead cells (cells with disrupted outer membranes).
Evaluation of apoptosis with automated cellular imaging
After staining, live cells were imaged with ImageXpress® Pico Automated Cell Imaging System using the 10X objective. Images were acquired at one site per well with DAPI, FITC and TRITC channels at 20, 500, and 50 ms exposure times, respectively. Figure 1 shows images of control cells and cells treated with 0.3 µM staurosporine and 10 µM mitomycin C.
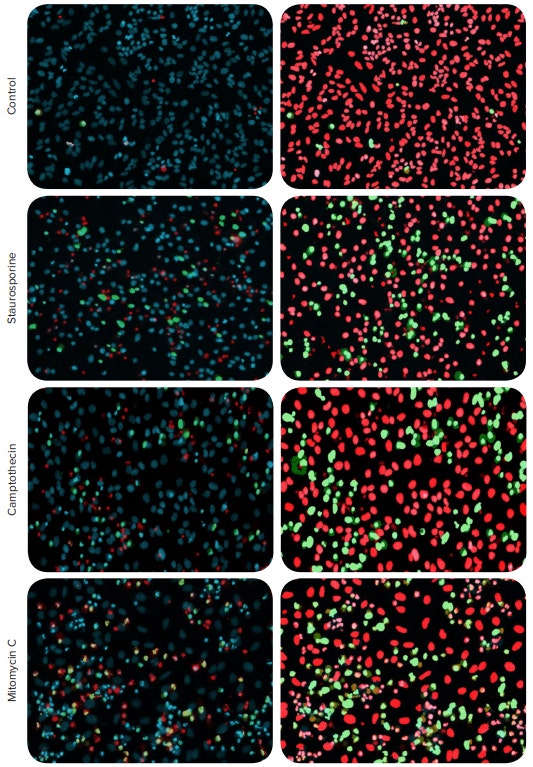
Figure 1. Detection of apoptosis using automated cell imaging. HeLa cells were treated with anti-cancer compounds. The compound concentrations for the representative images are 0.3 µM staurosporine, 66 µM of camptothecin, and 22 µM mitomycin C. The cells were stained with EarlyTox Caspase-3/7 NucView 488 Assay Kit in combination with Hoechst nuclear dye and Ethidium Homodimer III. Composite images shown on the left panel: nuclear stain Hoechst indicated in blue, apoptotic nuclei shown in green, dead cells nuclei shown in red. Apoptosis analysis overlay masks shown on the right: apoptotic nuclei (green) and non-apoptotic nuclei (red).
The Hoechst nuclear stain is shown in blue, apoptotic nuclei indicated by green, and nuclei of dead cells labeled in red. Using the Apoptosis analysis protocol in the CellReporterXpress Image Acquisition and Analysis Software together with image acquisition on the fly, the number and percentage of apoptotic cells, as indicated by the caspase staining, were evaluated. In addition, the Ethidium Homodimer stain can also be used to detect number and percentages of dead cells, and the Hoechst stain was used to define total cells. The resulting concentration response curves for induction of apoptosis and cell death along with the measured EC50 values are shown in Figure 2. Notably, the EC50 for kinase inhibitor staurosporine, a known potent inducer of apoptosis, was approximately 100 times lower than that of mitomycin C , which induces an anticancer effect through a different cellular mechanism (bioreductive alkylation)
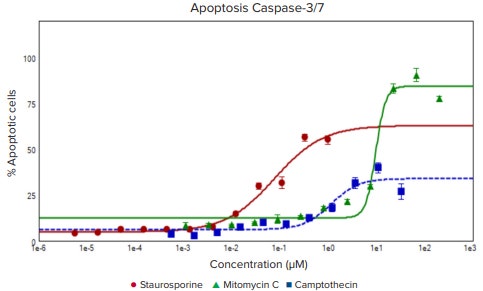
Figure 2. Concentration-responses for three anti-cancer compounds. The percentage of apoptotic cells: staurosporine (red, EC50 0.076 µM), mitomycin C (green, EC50 9.669 µM), and camptothecin (blue,EC50 1.14 µM).
Conclusion
The EarlyTox assay kit is useful for studying the biology of diseases, evaluating anti-cancer drugs, or monitoring cell health. It demonstrates the utility of the ImageXpressPico system and CellReporterXpress software in the evaluation and quantitation of apoptosis
References
- Green, Douglas (2011). Means to an End: Apoptosis and other Cell Death Mechanisms. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. ISBN 978-0-87969-888-1.
- Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2008). Molecular Biology of the Cell. (5th ed.). Chapter 18. Apoptosis: Programmed Cell Death Eliminates Unwanted Cells. Garland Science. p. 1115. ISBN 978-0-8153-4105-5.
- Karam, Jose A. (2009). Apoptosis in Carcinogenesis and Chemotherapy. Netherlands: Springer. ISBN 978-1-4020-9597-9.
- Verweij J1, Pinedo HM. Mitomycin C: mechanism of action, usefulness and limitations. Anticancer Drugs. 1990 Oct;1(1):5-13.